P1 – Self-assembly
Self-assembly of hierarchically structured surfaces and complexes can be a fascinatingly efficient process and we are only starting to grasp the molecular mechanism of these out-of-equilibrium systems. Here, we focus on assembly of proteins driven by surface anchored synthetic molecular rotary motors and synthetic as well as natural supramolecular assemblies. For the motors we look at their dynamic and adaptive functions, and how these structures are capable of directing cellular morphology and differentiation. Furthermore, we look at the self-assembly of viral nanoparticles and their engineered counterparts: synthetic regularly ordered assemblies. For both we scrutinize assembly kinetics, and for the viruses we also dive into the function of specific anchors, such as packaging signals, that direct and expedite self-assembly.
PhD-project 1a: Self-assembly of surface proteins steered by molecular motors: computational modelling
PhD student: Petteri Vainikka
Supervisors: Marrink / Browne
Aim:
Our goal is to promote new avenues of investigation through molecular simulation, and provide molecular-level details to existing topics. The main focus of our work lies in the biomolecule – surface interactions, which give rise to larger scale phenomena observed in the systems studied by this consortium.
Method:
We employ classical equilibrium and non-equilibrium molecular dynamics on both atomistic and coarse-grained level. Atomistic simulations are performed with well established force fields such as CHARMM, whereas the coarse-grained simulations are performed with Martini 3, which is developed in-house. Combined, this approach allows us to observe phenomena in temporal and spatial resolutions which are currently either extremely challenging or impossible to capture using experimental methods.
PhD-project 1b: Self-assembly of surface proteins steered by molecular motors: experiments
PhD student: Daniel Döllerer
Supervisors: Feringa / Van Rijn
Aim:
To achieve higher order cellular complexity at 2D interfaces and 3D matrices using dynamic control provided by surface confined unidirectional rotary molecular motors.
Method:
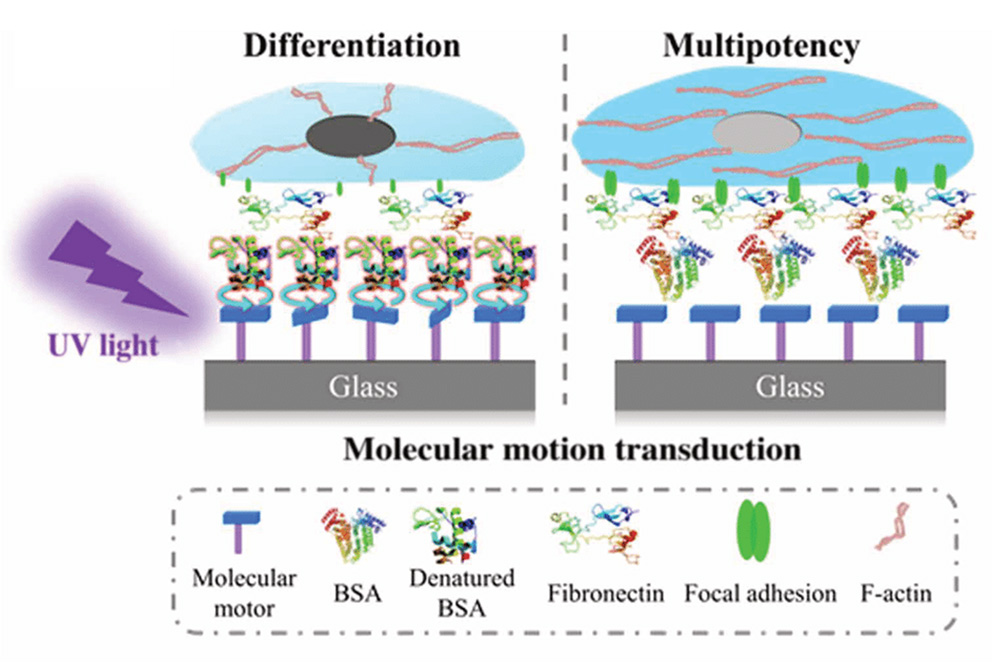
Molecular motors based on overcrowded alkenes can be beside other possible applications, grafted via different methods, such as click chemistry, amide bond formation or electrostatic interactions onto various surfaces, e.g. gold, glass, or nanoparticles. These fixed motors are then capable to convert energy gained by irradiation into controlled motion. As a consequence of this operation, a system can be driven out-of-equilibrium with molecular precision and thus influence the fate of stem cells, promoting differentiation.
However, most tissues have a 3D morphology and cell distribution, which makes it ultimately appreciable to move from 2D interfaces into 3D matrices as a growing surface. As a consequence, molecular motors attached to fiber spun meshes orchestrate more complexity and therefore a higher degree of biomimicry is obtained. Additionally, the use of photomasks generates light-induced patterns and can then direct cell adhesion. This selective adhesion results in a plethora of applications to generate double or triple cell type compositions paving a way to create 3D tissue like structures.
PhD-project 1c: Signal-driven self-assembly of viral particles
PhD student: Chris van Ewijk
Supervisors: W.H. Roos / P.R. Onck
Aim:
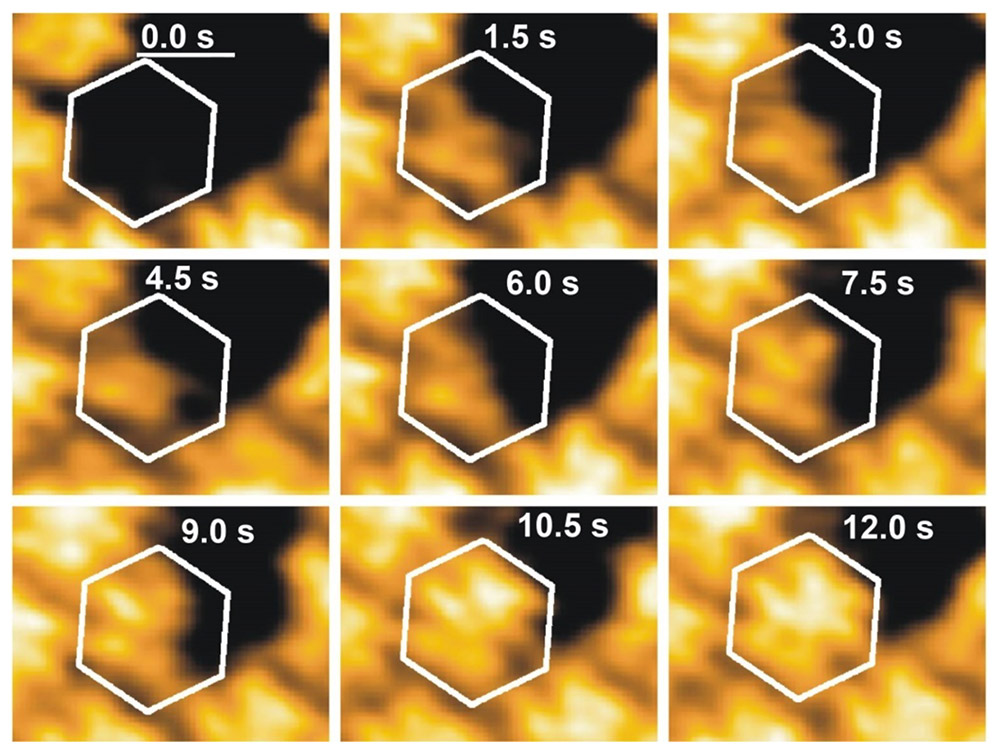
We aim to discover the self-assembly kinetics of viral and synthetic nanostructures and to scrutinize the specific interactions that direct the self-assembly process.
Method:
To visualize these dynamic and fast processes in real time we make use of High Speed Atomic Force Microscopy (HS-AFM). Here a small cantilever (≤10µm) is oscillated at high frequency (≥500kHz) and scanned over the sample. By detecting the deflection of the cantilever the height profile of the sample can be determined with up to 10 frames per second. This allows us to follow the dynamic self-assembly processes and to deduce kinetic parameters from it.