P3 – Self-replication
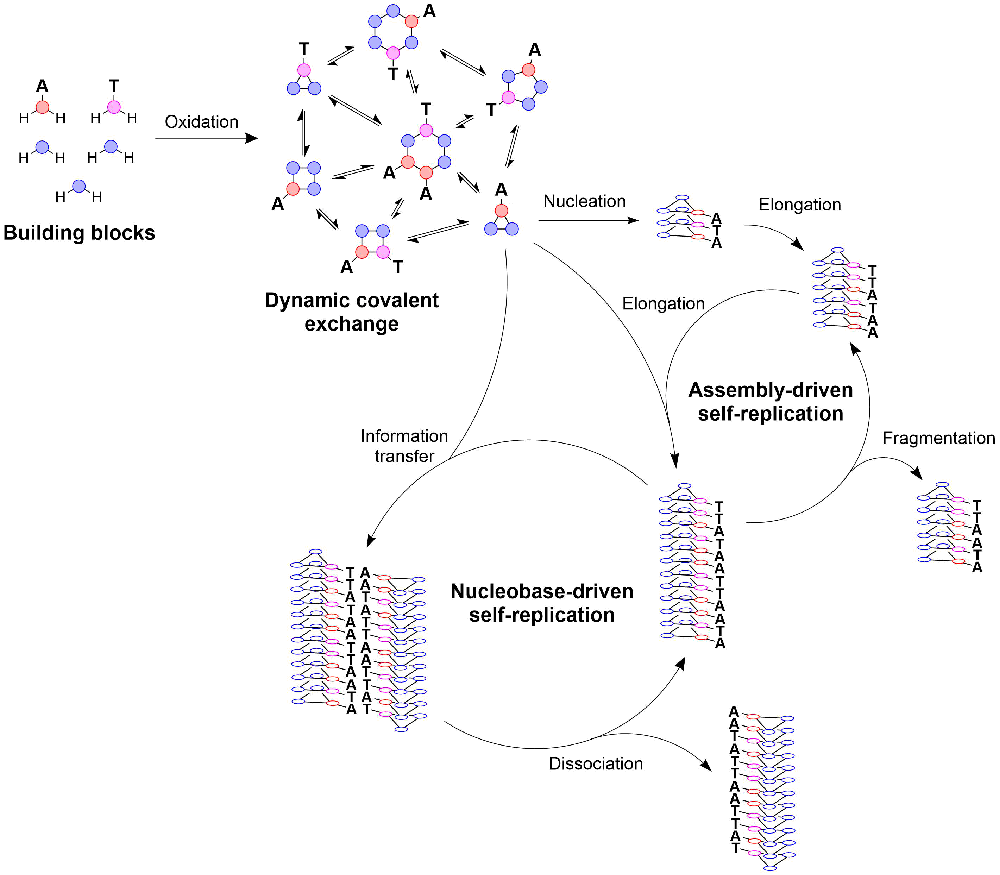
How systems capable of replicating nucleobase sequences have emerged remains shrouded in mystery. Using a combination of coarse-grained simulations and experimental work we aim to explore a new pathway to nucleobase-containing replicators which, unlike previous approaches, starts with simple building blocks containing only a single nucleobase. Preliminary results have confirmed that, through a process of auto-catalytic assembly-driven self-replication, stacks of rings can be obtained that display nucleobases on their periphery. The aim is now to devise strategies that result in the transfer of the information contained in the sequence of nucleobases along the stack, from existing stacks to newly forming ones. Subjecting these out-of-equilibrium systems to a regime in which replication and replicator destruction occur simultaneously, while replicators can mutate at the same time, should result in Darwinian-type evolution of the nucleobase-containing replicators.
PhD-project 3a: Nucleobase-driven synthetic self-replication: experiments
PhD student: Marc Geerts
Supervisors: Otto / Van der Wel
Aim/method:
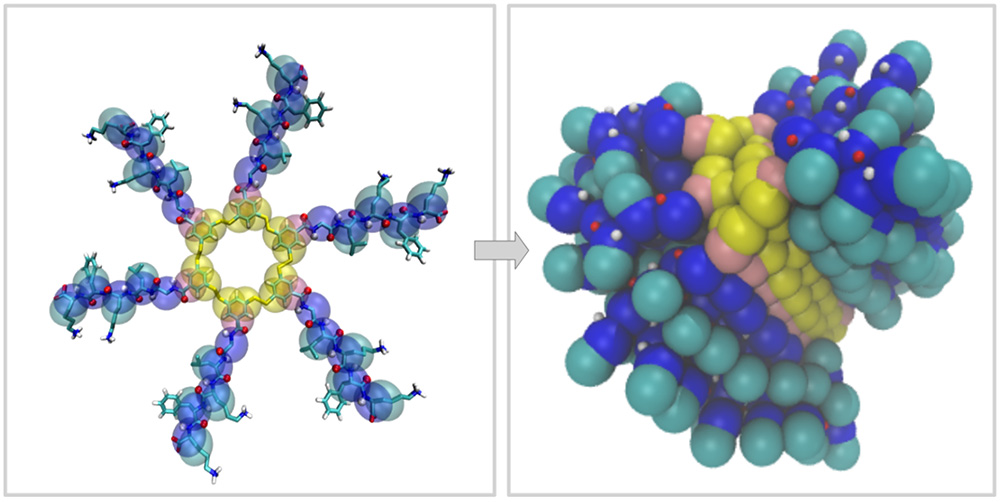
The aim is to synthesise new building blocks consisting of a dimercaptobenzene connected to a nucleobase through a linker. Oxidation of the thiols to disulfides results in the formation of rings composed of multiple building blocks, which can subsequently self-assemble into stacks capable of assembly-driven self-replication through stack-elongation and fragmentation. By mixing different building blocks and varying them in terms of length, rigidity, and hydrophobicity, we aim to identify what design factors are critical for obtaining stacks which can transfer the information contained within the nucleobase sequence to newly forming stacks through base-pairing. The composition of rings within a stack will be determined using ultraperformance liquid chromatography-mass spectrometry, whereas the stack-structure and inter-stack interactions will be characterized using spectroscopic and microscopic techniques. These observations will be combined with and compared to computational models of this system. See project 3b.
PhD-project 3b: Nucleobase-driven synthetic self-replication: computational modelling
PhD student: Mark van der Klok
Supervisors: Onck / Otto
Aim:
The aim of this project is to assist the development of new nucleobase-containing self-replicators, by computationally studying their structure, nucleobase accessibility and self-assembly. By performing a detailed analysis we try to understand the underlying mechanisms and interactions and to correlate simulation readouts with replication propensity.
Method:
We use molecular dynamics simulations to study the self-replicators, their fiber structure and their self-assembly process. To study fiber structures at shorter length scales, we use simulations at the atomistic scale. Additionally, we work on larger-scale simulations using our newly-developed 4-bead-per-aminoacid (4BPA) coarse-grained force field.